Summary
Clan
This family is a member of clan (CL00106), which contains the following 10 members:
CsrB CsrC McaS PrrB_RsmZ RsmV RsmW rsmX RsmY TwoAYGGAY Ysr186_sR026_CsrCWikipedia annotation Edit Wikipedia article
The Rfam group coordinates the annotation of Rfam families in Wikipedia. This family is described by a Wikipedia entry CsrB/RsmB RNA family. More...
This page is based on a Wikipedia article. The text is available under the Creative Commons Attribution/Share-Alike License.
Sequences
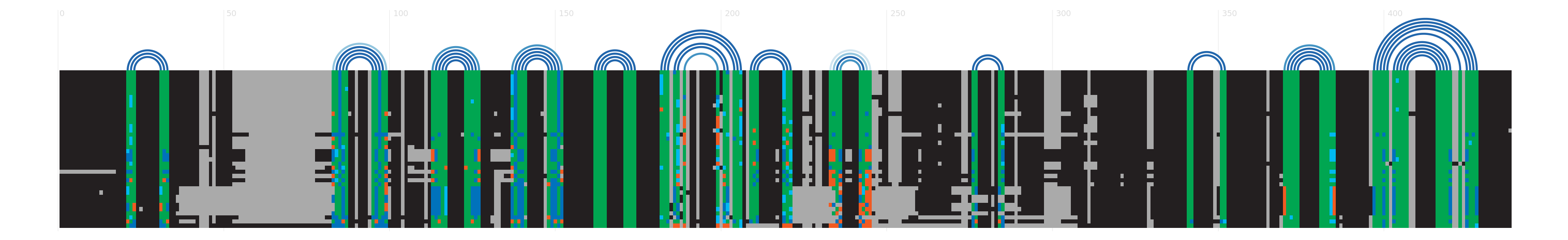
Alignment
There are various ways to view or download the seed alignments that we store. You can use a sequence viewer to look at them, or you can look at a plain text version of the sequence in a variety of different formats. More...
View options
You can view Rfam seed alignments in your browser in various ways. Choose the viewer that you want to use and click the "View" button to show the alignment in a pop-up window.
Formatting options
You can view or download Rfam seed alignments in several formats. Check either the "download" button, to save the formatted alignment, or "view", to see it in your browser window, and click "Generate".
Download
Download a gzip-compressed, Stockholm-format file containing the seed alignment for this family. You may find RALEE useful when viewing sequence alignments.
Submit a new alignment
We're happy receive updated seed alignments for new or existing families. Submit your new alignment and we'll take a look.
Secondary structure
Species distribution
Sunburst controls
HideWeight segments by...
Change the size of the sunburst
Colour assignments
 Archea
Archea
|
 Eukaryota
Eukaryota
|
 Bacteria
Bacteria
|
 Other sequences
Other sequences
|
 Viruses
Viruses
|
 Unclassified
Unclassified
|
 Viroids
Viroids
|
 Unclassified sequence
Unclassified sequence
|
Selections
Click on a node to select that node and its sub-tree.
Clear selection
This visualisation provides a simple graphical representation of the distribution of this family across species. You can find the original interactive tree in the adjacent tab. More...
Tree controls
HideThe tree shows the occurrence of this RNA across different species. More...
Loading...
Please note: for large trees this can take some time. While the tree is loading, you can safely switch away from this tab but if you browse away from the family page entirely, the tree will not be loaded.
Trees
This page displays the predicted phylogenetic tree for the alignment. More...
Note: You can also download the data file for the seed tree.
Motif matches
There are 7 motifs which match this family.
This section shows the Rfam motifs that match sequences within the seed alignment of this family. Users should be aware that the motifs are structural constructs and do not necessarily conform to taxonomic boundaries in the way that Rfam families do. More...
| Original order | Motif Accession | Motif Description | Number of Hits | Fraction of Hits | Sum of Bits | Image |
|---|---|---|---|---|---|---|
| 7 | RM00005 | CsrA/RsmA binding motif | 38 | 1.000 | 4480.4 |
|
| 7 | RM00007 | Splicing domain V | 19 | 0.500 | 202.5 |
|
| 7 | RM00008 | GNRA tetraloop | 23 | 0.605 | 261.7 |
|
| 7 | RM00022 | Rho independent terminator 1 | 31 | 0.816 | 440.7 |
|
| 7 | RM00023 | Rho independent terminator 2 | 35 | 0.921 | 660.3 |
|
| 7 | RM00029 | UNCG tetraloop | 19 | 0.500 | 157.5 |
|
| 7 | RM00030 | U-turn motif | 8 | 0.211 | 80.8 |
|
References
This section shows the database cross-references that we have for this Rfam family.
Literature references
-
Liu MY, Gui G, Wei B, Preston JF 3rd, Oakford L, Yuksel U, Giedroc DP, Romeo T J Biol Chem 1997;272:17502-17510. The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. PUBMED:97362239
-
Cui Y, Chatterjee A, Liu Y, Dumenyo CK, Chatterjee AK J Bacteriol 1995;177:5108-5115. Identification of a global repressor gene, rsmA, of Erwinia carotovora subsp. carotovora that controls extracellular enzymes, N-(3-oxohexanoyl)-L-homoserine lactone, and pathogenicity in soft-rotting Erwinia spp. PUBMED:7665490
-
Righetti F, Nuss AM, Twittenhoff C, Beele S, Urban K, Will S, Bernhart SH, Stadler PF, Dersch P, Narberhaus F Proc Natl Acad Sci U S A. 2016;113:7237-7242. Temperature-responsive in vitro RNA structurome of Yersinia pseudotuberculosis. PUBMED:27298343
-
Heroven AK, Sest M, Pisano F, Scheb-Wetzel M, Steinmann R, Bohme K, Klein J, Munch R, Schomburg D, Dersch P Front Cell Infect Microbiol. 2012;2:158. Crp induces switching of the CsrB and CsrC RNAs in Yersinia pseudotuberculosis and links nutritional status to virulence. PUBMED:23251905
-
Mei L, Xu S, Lu P, Lin H, Guo Y, Wang Y PLoS One. 2017;12:e0187492. CsrB, a noncoding regulatory RNA, is required for BarA-dependent expression of biocontrol traits in Rahnella aquatilis HX2. PUBMED:29091941
-
Yang S, Peng Q, Zhang Q, Yi X, Choi CJ, Reedy RM, Charkowski AO, Yang CH Mol Plant Microbe Interact. 2008;21:133-142. Dynamic regulation of GacA in type III secretion, pectinase gene expression, pellicle formation, and pathogenicity of Dickeya dadantii (Erwinia chrysanthemi 3937). PUBMED:18052890
-
Cui Y, Chatterjee A, Yang H, Chatterjee AK J Bacteriol. 2008;190:4610-4623. Regulatory network controlling extracellular proteins in Erwinia carotovora subsp. carotovora: FlhDC, the master regulator of flagellar genes, activates rsmB regulatory RNA production by affecting gacA and hexA (lrhA) expression. PUBMED:18441056
-
Yuan X, Khokhani D, Wu X, Yang F, Biener G, Koestler BJ, Raicu V, He C, Waters CM, Sundin GW, Tian F, Yang CH Environ Microbiol. 2015;17:4745-4763. Cross-talk between a regulatory small RNA, cyclic-di-GMP signalling and flagellar regulator FlhDC for virulence and bacterial behaviours. PUBMED:26462993
External database links
| Gene Ontology: | GO:0005515 (protein binding); |
| Sequence Ontology: | SO:0000377 (CsrB_RsmB_RNA); |
Curation and family details
This section shows the detailed information about the Rfam family. We're happy to receive updated or improved alignments for new or existing families. Submit your new alignment and we'll take a look.
Curation
| Seed source | Bateman A | ||||||
| Structure source | Predicted; PFOLD; Bateman A; Gardner PP; | ||||||
| Type | Gene; sRNA; | ||||||
| Author |
Bateman A ,
Gardner PP ,
Gardner PP
|
||||||
| Alignment details |
|
Model information
| Build commands |
cmbuild -F CM SEED
cmcalibrate --mpi CM
|
| Search command |
cmsearch --cpu 4 --verbose --nohmmonly -T 19.98 -Z 2958934 CM SEQDB
|
| Gathering cutoff | 71.0 |
| Trusted cutoff | 71.0 |
| Noise cutoff | 70.9 |
| Covariance model | Download |
 Loading...
Loading...