Summary
Clan
This family is a member of clan (CL00007), which contains the following 2 members:
U4 U4atacWikipedia annotation Edit Wikipedia article
The Rfam group coordinates the annotation of Rfam families in Wikipedia. This family is described by a Wikipedia entry U4 spliceosomal RNA. More...
This page is based on a Wikipedia article. The text is available under the Creative Commons Attribution/Share-Alike License.
Sequences
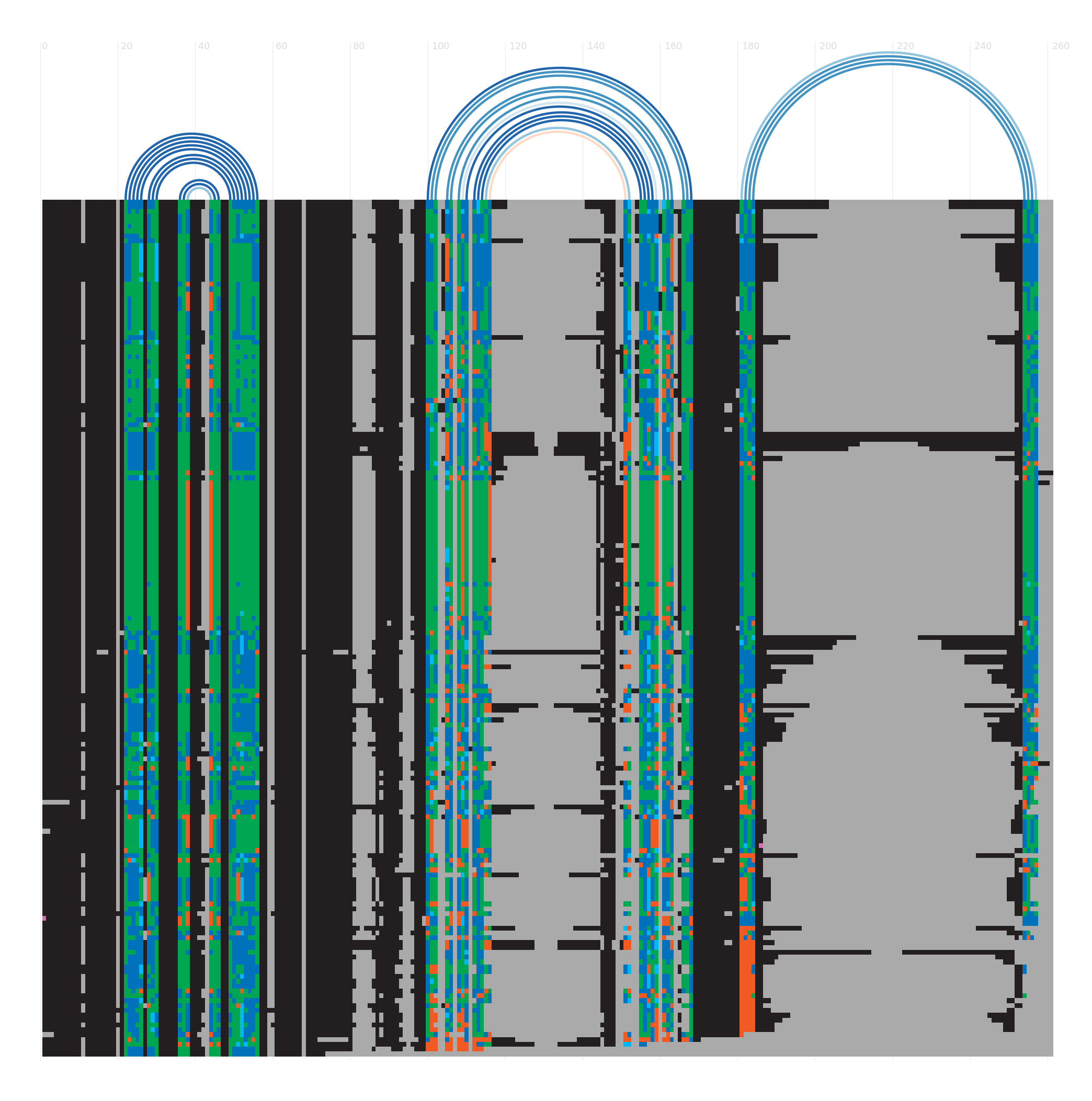
Alignment
There are various ways to view or download the seed alignments that we store. You can use a sequence viewer to look at them, or you can look at a plain text version of the sequence in a variety of different formats. More...
View options
You can view Rfam seed alignments in your browser in various ways. Choose the viewer that you want to use and click the "View" button to show the alignment in a pop-up window.
Formatting options
You can view or download Rfam seed alignments in several formats. Check either the "download" button, to save the formatted alignment, or "view", to see it in your browser window, and click "Generate".
Download
Download a gzip-compressed, Stockholm-format file containing the seed alignment for this family. You may find RALEE useful when viewing sequence alignments.
Submit a new alignment
We're happy receive updated seed alignments for new or existing families. Submit your new alignment and we'll take a look.
Secondary structure
Species distribution
Sunburst controls
HideWeight segments by...
Change the size of the sunburst
Colour assignments
 Archea
Archea
|
 Eukaryota
Eukaryota
|
 Bacteria
Bacteria
|
 Other sequences
Other sequences
|
 Viruses
Viruses
|
 Unclassified
Unclassified
|
 Viroids
Viroids
|
 Unclassified sequence
Unclassified sequence
|
Selections
Click on a node to select that node and its sub-tree.
Clear selection
This visualisation provides a simple graphical representation of the distribution of this family across species. You can find the original interactive tree in the adjacent tab. More...
Tree controls
HideThe tree shows the occurrence of this RNA across different species. More...
Loading...
Please note: for large trees this can take some time. While the tree is loading, you can safely switch away from this tab but if you browse away from the family page entirely, the tree will not be loaded.
Trees
This page displays the predicted phylogenetic tree for the alignment. More...
Note: You can also download the data file for the seed tree.
Structures
For those sequences which have a structure in the Protein DataBank, we generate a mapping between EMBL, PDB and Rfam coordinate systems. The table below shows the structures on which the U4 family has been found.
Loading structure mapping...
Motif matches
There are 1 motifs which match this family.
This section shows the Rfam motifs that match sequences within the seed alignment of this family. Users should be aware that the motifs are structural constructs and do not necessarily conform to taxonomic boundaries in the way that Rfam families do. More...
| Original order | Motif Accession | Motif Description | Number of Hits | Fraction of Hits | Sum of Bits | Image |
|---|---|---|---|---|---|---|
| 7 | RM00029 | UNCG tetraloop | 38 | 0.215 | 533.7 |
|
References
This section shows the database cross-references that we have for this Rfam family.
Literature references
-
Zwieb C Nucleic Acids Res 1997;25:102-103. The uRNA database. PUBMED:9016512
-
Vidovic I, Nottrott S, Hartmuth K, Luhrmann R, Ficner R Mol Cell 2000;6:1331-1342. Crystal structure of the spliceosomal 15.5kD protein bound to a U4 snRNA fragment. PUBMED:11163207
-
Raghunathan PL, Guthrie C Science 1998;279:857-860. A spliceosomal recycling factor that reanneals U4 and U6 small nuclear ribonucleoprotein particles. PUBMED:9452384
-
Thomas J, Lea K, Zucker-Aprison E, Blumenthal T Nucleic Acids Res 1990;18:2633-2642. The spliceosomal snRNAs of Caenorhabditis elegans. PUBMED:2339054
-
Cornilescu G, Didychuk AL, Rodgers ML, Michael LA, Burke JE, Montemayor EJ, Hoskins AA, Butcher SE J Mol Biol. 2016;428:777-789. Structural Analysis of Multi-Helical RNAs by NMR-SAXS/WAXS: Application to the U4/U6 di-snRNA. PUBMED:26655855
-
Wan R, Yan C, Bai R, Wang L, Huang M, Wong CC, Shi Y Science. 2016;351:466-475. The 3.8 A structure of the U4/U6.U5 tri-snRNP: Insights into spliceosome assembly and catalysis. PUBMED:26743623
-
Nguyen TH, Galej WP, Bai XC, Savva CG, Newman AJ, Scheres SH, Nagai K Nature. 2015;523:47-52. The architecture of the spliceosomal U4/U6.U5 tri-snRNP. PUBMED:26106855
-
Nguyen THD, Galej WP, Bai XC, Oubridge C, Newman AJ, Scheres SHW, Nagai K Nature. 2016;530:298-302. Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 A resolution. PUBMED:26829225
-
Plaschka C, Lin PC, Nagai K Nature. 2017;546:617-621. Structure of a pre-catalytic spliceosome. PUBMED:28530653
-
Bertram K, Agafonov DE, Dybkov O, Haselbach D, Leelaram MN, Will CL, Urlaub H, Kastner B, Luhrmann R, Stark H Cell. 2017;170:701-713. Cryo-EM Structure of a Pre-catalytic Human Spliceosome Primed for Activation. PUBMED:28781166
-
Bai R, Wan R, Yan C, Lei J, Shi Y Science. 2018;360:1423-1429. Structures of the fully assembled Saccharomyces cerevisiae spliceosome before activation. PUBMED:29794219
-
Zhan X, Yan C, Zhang X, Lei J, Shi Y Cell Res. 2018;28:1129-1140. Structures of the human pre-catalytic spliceosome and its precursor spliceosome. PUBMED:30315277
-
Charenton C, Wilkinson ME, Nagai K Science. 2019;364:362-367. Mechanism of 5' splice site transfer for human spliceosome activation. PUBMED:30975767
External database links
| Gene Ontology: | GO:0000244 (spliceosomal tri-snRNP complex assembly); GO:0000353 (formation of quadruple SL/U4/U5/U6 snRNP); GO:0017070 (U6 snRNA binding); GO:0005687 (U4 snRNP); GO:0046540 (U4/U6 x U5 tri-snRNP complex); |
| Sequence Ontology: | SO:0000393 (U4_snRNA); |
Curation and family details
This section shows the detailed information about the Rfam family. We're happy to receive updated or improved alignments for new or existing families. Submit your new alignment and we'll take a look.
Curation
| Seed source | Zwieb C, The uRNA database, PMID:9016512 | ||||||
| Structure source | Published; PMID:2339054; Griffiths-Jones SR | ||||||
| Type | Gene; snRNA; splicing; | ||||||
| Author |
Griffiths-Jones SR ,
Ontiveros-Palacios N ,
Ontiveros-Palacios N
|
||||||
| Alignment details |
|
Model information
| Build commands |
cmbuild -F CM SEED
cmcalibrate --mpi CM
|
| Search command |
cmsearch --cpu 4 --verbose --nohmmonly -T 30.00 -Z 2958934 CM SEQDB
|
| Gathering cutoff | 46.0 |
| Trusted cutoff | 46.0 |
| Noise cutoff | 45.9 |
| Covariance model | Download |
 Loading...
Loading...